Stout camphor tree is one of the many remarkable plant species endemic to Taiwan. Conservation of stout camphor tree has become especially urgent as poachers have been illegally taking trees for cultivation of a specialized fungus used in traditional medicine and reported to have cancer-fighting agents. Now a team of scientists lead by researchers at Academia Sinica’s Biodiversity Research Center in Taiwan has decoded a complete genome sequence for stout camphor tree. The article is published on Nature Plants on January 11, 2019.
In addition to its utility in the development of new cancer treatments, the stout camphor trees are a culturally important part of the broadleaf evergreen forests in Taiwan. Trees can live for more than one thousand years with trunk diameters reaching more than over 2 meters! Aromatic and decay-resistant stout camphor tree wood is used for high-end furniture and art production. The high value of stout camphor tree together with intensive deforestation over the past half century, and poor seed germination have contributed to the loss and fragmentation of natural populations.
Stout camphor tree along with magnolia, tulip tree, custard apple and nutmeg are “magnoliids”, a group comprising about 6% of all flowering plant diversity. The relationship of magnoliids to other diverse plant groups (moncots and eudicots) has been uncertain despite years of investigation by evolutionary botanists. The new stout camphor tree genome sequence has enabled improved researchers to pinpoint the position of magnoliids within the flowering plant tree of life while at the same time elucidating the complex history of magnoliid genome evolution. Interestingly, analyses revealed two rounds of ancient genome duplications since the magnoliids diverged from other flowering plant lineages approximately 150 million years ago. At the same time, genes involved in the production of diverse terpenoid compounds multiplied in the ancestral stout camphor tree genome, contributing to its complex aromatic properties and wood chemistry.
Additional analyses were able to detect evidence for steady declines in stout camphor tree populations over the last million years. Corresponding author Dr. Isheng J. Tsai hypothesizes that recent losses of stout camphor tree populations associated with over harvesting were predated by longer term declines driven by the complex geological history of Taiwan. Dr. Tsai also emphasizes that their published reference genome will be useful for current efforts to monitor and maintain genetic diversity in remaining for stout camphor tree populations.
First author, Dr. Shu-Miaw Chaw, sums up the team’s effort stating that “the availability of a complete stout camphor tree genome sequence establishes a valuable genomic foundation that will shed light on genetic diversity within this culturally and economically important broadleaved forest species, and at the same time help researchers gain a better understanding of magnoliid and flowering plant genome evolution and diversification.”
The corresponding authors are Shu-Miaw Chaw, Distinguished Research Fellow, and Isheng J. Tsai, Assistant Research Fellow, of Biodiversity Research Center, Academia Sinica. This study is supported by Investigator Award and Thematic Award to SMC, and by a Career Award to IJT from Academia Sinica. Dr. James H. Leebens-Mack, professor of Department of Plant Biology, University of Georgia, contributed transcriptomic data and interpretation of analytical results. This article was published on the issue of January 9, 2019, Nature Plants (full article is available at https://rdcu.be/bfUNQ ). A commentary reports the importance of this article along with the Chinese tulip tree genome appears in the same issue1.
1 Nuclear genomes of two magnoliids: https://www.nature.com/articles/s41477-018-0344-1

▲ Fig. 1︳A wild stout camphor tree inside a dense broadleaved forest near Tengzi. Photo was taken on December 5, 2018 with assistance from administrator of Forestry Bureau, Pingtung District.
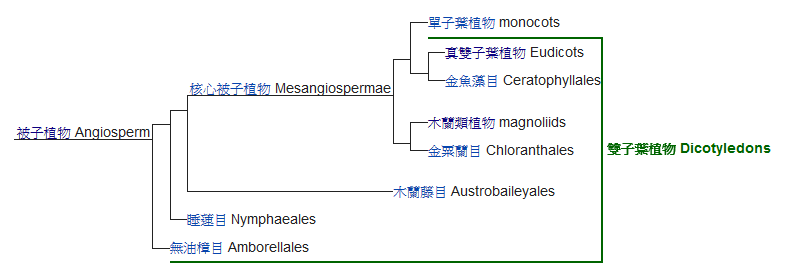
▲ Fig. 2︳ A species tree on the basis of 211 single copy orthologues from 13 plant species. Gene family expansion and contraction are denoted in numbers next to plus and minus signs, respectively. Green number in brackets denote divergence time estimates. Time of divergence between magnoliids and eudicots to be 136.0–209.4 million years ago (Ma)
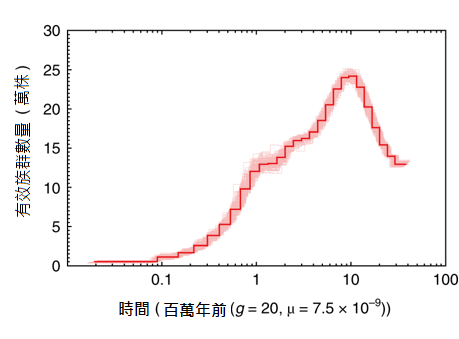
▲Fig. 3︳Stout camphor tree population size dynamics from the past 9 million years ago to present. The history of effective population size was inferred using the PSMC method. Ma: Million years ago.
