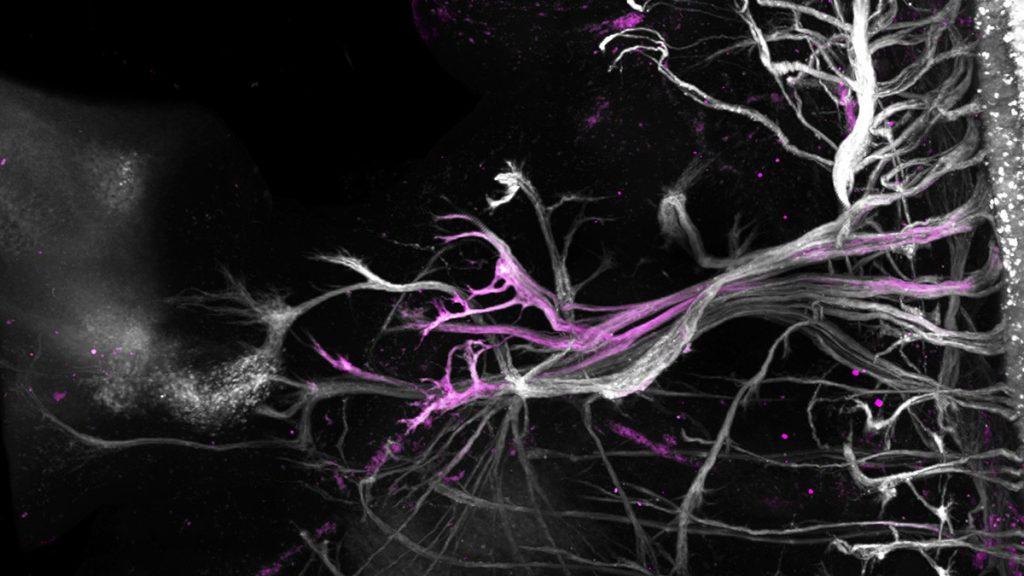
A tour de force study by Dr. Jun-An Chen’s group at the Institute of Molecular Biology, Academia Sinica, has uncovered the diversity of spinal motor neurons in mouse embryos through single-cell RNA sequencing. Their study scrutinizing embryonic motor neurons at single-cell resolution reports rare cell types and their genetic markers, explains the transcriptomic relationships between motor neuron groups, and compares cell type diversity across different species. The group sequenced 10,000 mouse embryonic spinal motor neurons, among which new types were annotated. This remarkable effort provides a single-cell atlas of spinal motor neurons and serves as an invaluable resource for the neuroscience community, laying the foundation for establishing the mysterious vulnerability of motor neurons to some neurodegenerative diseases, such as spinal muscular atrophy and amyotrophic lateral sclerosis. The study “Single-cell transcriptomic analysis reveals diversity within mammalian spinal motor neurons” has been published in Nature Communications.
Decades of effort by multiple research groups has elegantly characterized motor neuron diversity in the spinal cord by means of retrograde-labeling the different muscle groups and tracing their innervating motor neurons. Traditionally, motor neurons are categorized based on cell body position and this innervation pattern. Upon their genetic ablation, motor neurons manifest identity and positional alterations and axonal targeting/arborization deficits, providing evidence that intrinsic and extrinsic mechanisms shape motor neuron diversity and functions. Based on this observation, this study led by Dr. Chen molecularly characterized spinal motor neuron diversity unbiasedly and uncovered rare and new segment-specific motor neurons in vivo. Then, focusing on known MN subtypes, Dr. Chen’s group subclustered cells displaying limb-innervating identity (LMCs) into 26 clusters and surprisingly observed diverse expression of neuropeptides in the brachial LMC motor neurons and between the brachial and lumbar LMCs. That neuropeptides exert a function during embryonic development has not been reported extensively. They speculate that neuropeptides in motor neurons serve important signaling roles, such as in precise axon targeting, modulating spinal circuit formation, or even facilitating divergent activity patterns, all representing fascinating avenues for future study.
Moreover, their study highlights the underappreciated diversity of motor neurons in the body trunk. They have unveiled three molecularly-distinct subpopulations among this group of motor neurons, each represented by differentially expressed transcription factors and divergent innervation targets. Even though trunk motor neurons generally control axial muscle contraction, they execute distinct motor behaviors between aquatic and land vertebrates. How trunk motor neurons evolved during the dramatic evolutionary transition from water to land is the next fundamental question being tackled by Dr. Chen’s group. This study provides a rich transcriptomic resource of motor neuron subtypes, reveals the molecular logic underlying the developmental diversity of motor neurons, and illuminates subtype diversity across different species to provide insights into the shift in motor behavior as chordates adapted from water to land.
This study is a collaboration between Academia Sinica in Taiwan, University of California Irvine (UCI) in the USA, and Institut Du Fer À Moulin in France. The primary authors of the article are Ee Shan Liau, a TIGP student in Academia Sinica, and Dr. Suoqin Jin from UCI (now an associate professor at Wuhan University in China). Academia Sinica and the National Science and Technology Council in Taiwan funded the study.
The full research article can be viewed at the following link:
https://www.nature.com/articles/s41467-022-35574-x