Completion of Chromosome-level Assembly, Genetic and Physical Maps of Orchid Phalaenopsis aphrodite Genome to Advance Orchid Industry Molecular Breeding
Distinguished Research Fellow from the Agricultural Biotechnology Research Center, Dr. Ming-Che Shih and colleagues have recently compiled a quality reference genome and genetic map of the orchid Phalaenopsis aphrodite that provides new insights into the orchid genome architecture and will serve as a crucial resource for orchid breeding, orchid species conservation and comparative genomic studies. The work was published in the Plant Biotechnology Journal on April 28.

Phalaenopsis Aphrodite
Taiwan’s orchid export industry has an estimated annual export value of more than US$130 million. In order to meet and stimulate consumer demand in the market, the industry urgently needs to introduce new varieties of orchids; however, the growth cycle of orchids is as long as two to three years. The traditional breeding method, is time-consuming, energy-consuming, and has low success rates. Therefore, the industry has moved toward using molecular breeding techniques to generate desirable new varieties. Basic information about Taiwan orchid genetics can identify important molecular markers, establish high-density genetic linkage maps, and accurately design a new generation of orchids with attractive genetic traits.
The orchid species Phalaenopsis aphrodite is native to Taiwan and has a beautiful flower and stalk shape making it a major breeding parent of many commercial orchid hybrids.
Chromosome-level assembly and genetic and physical mapping of the P. aphrodite genome provides new insights into species adaptation and will provide an unprecedented resource for genomics-assisted orchid breeding. It vastly improves on the data provided by a previously assembled draft orchid genome published in 2015, which was highly fragmented, had incorrectly assembled scaffolds, and provided neither genome architecture nor chromosome information.
The orchid genome information collected by the researchers over the years is publically available in the Orchidstra 2.0 database published by Academia Sinica at: http://orchidstra2.abrc.sinica.edu.tw/orchidstra2/index.php
The full article entitled “Chromosome-level assembly, genetic and physical mapping of Phalaenopsis aphrodite genome provides new insights into species adaptation and resources for orchid breeding” can be found at the Plant Biotechnology Journal website at: https://www.ncbi.nlm.nih.gov/pubmed/29704444
Supplementary data about P. aphrodite is available at: http://orchidstra2.abrc.sinica.edu.tw/orchidstra2/pagenome.php
The full list of authors is: Chao YT, Chen WC, Chen CY, Ho HY, Yeh CH, Kuo YT, Su CL, Yen SH, Hsueh HY, Yeh JH, Hsu HL, Tsai YH, Kuo TY, Chang SB, Chen KY, and Shih MC.
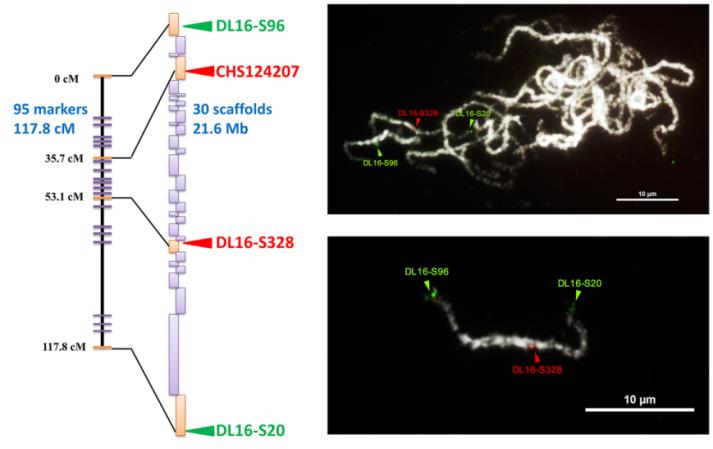
Fig.1. The integration of genetic map, genome assembly and FISH mapping.
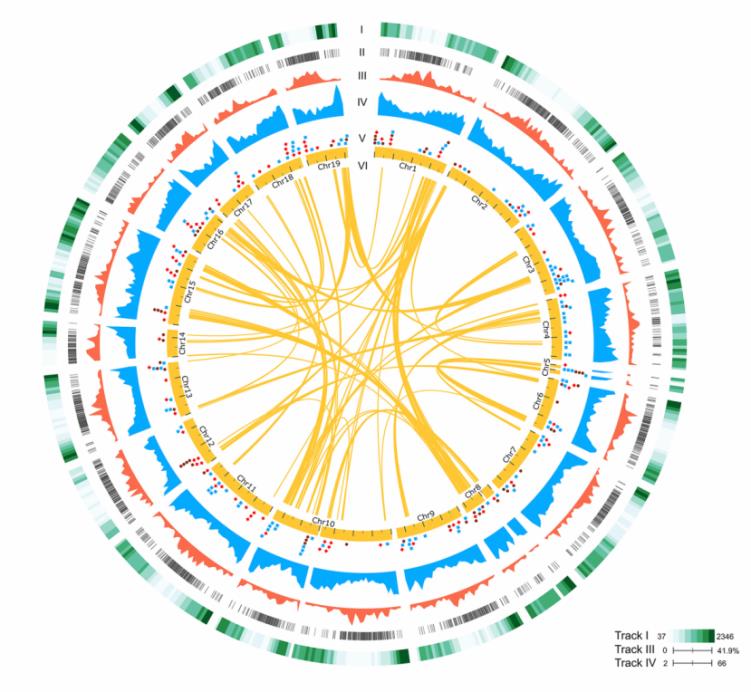
Fig.2. Genomic composition of P. aphrodite.
