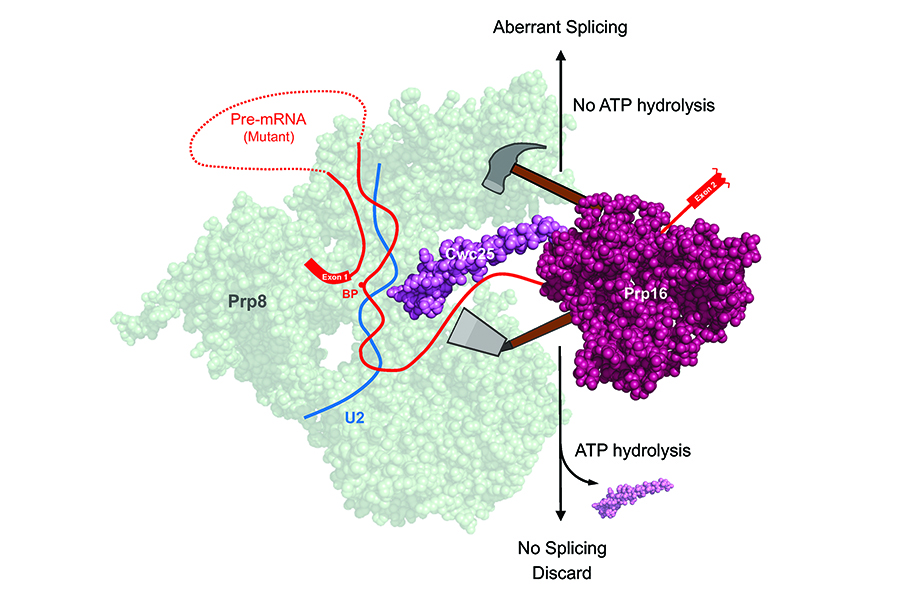
Most eukaryotic genes contain sequences that interrupt the genes’ continuity yet are not translated into proteins. These sequences are removed at the RNA level in a process termed RNA splicing, which occurs on a large ribonucleoprotein complex known as the spliceosome. The DExD/H-box RNA helicase Prp16 plays a pivotal role in the splicing process. Following the branching reaction, it utilizes its ATP-dependent functions to facilitate exon ligation by displacing step-one factors, Cwc25 and Yju2, from the spliceosome’s catalytic center, thus enabling the positioning of the 3’ splice site to the catalytic center. Additionally, Prp16 is well known for its role in proofreading the 5’ splice site and the branch site of pre-mRNA, a function that requires ATP. A team led by Distinguished Research Fellow Soo-Chen Cheng at the Institute of Molecular Biology, Academia Sinic has unveiled a paradoxical role for Prp16 in splicing of pre-mRNA with mutations at splice sites, resulting in a slower splicing reaction. In such cases, Prp16 exhibits an ATP-independent activity by stabilizing the binding of Cwc25, thereby enhancing the branching reaction and leading to aberrant selection of splice sites. This seemly contradictory function of Prp16 raises questions about its role in proofreading splice sites. Nevertheless, it underscores Prp16’s consistent role in propelling the splicing pathway forward. In normal pre-mRNA, Prp16 destabilizes Cwc25 to facilitate exon ligation, whereas for mutant pre-mRNA, it stabilizes Cwc25 to promotes the branching reaction. These discoveries change the current view of splicing fidelity control, leading to the paper’s recognition as Breakthrough Article by NAR.
Article link: https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkad861/7321994?login=true